Functional Screens to Identify Secreted Proteins
We have been developing and applying functional screens to identify genes encoding secreted proteins. The genes are identified based on the ultimate localization of the encoded protein, utilizing protein markers whose activities confirm targeting to the cell surface.
| Yeast Secretion Trap (YST) Screen |
Sucrose plate with no yeast colonies growing (negative control).
The YST screen can be used to test a specific cDNA of interest, or to screen a cDNA library. The screen uses invertase, an enzyme that hydrolyzes sucrose to generate glucose and fructose, as a reporter gene/protein and yeast (S. cerevisiae) as a host. If yeast cells are streaked onto plates containing sucrose as the sole carbon source, they must secrete invertase to break down the sucrose in order to grow, and so an invertase-deficient yeast mutant will be unable to grow.
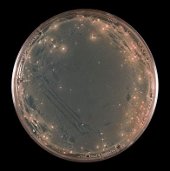
Sucrose plate showing yeast colonies transformed with the YST vector library.
This system can also identify plasma membrane (PM)-spanning or PM-anchored proteins if the orientation of the fusion protein is such that the invertase component is external to the cell. The resulting yeast transformants can then be isolated and the plasmids containing the heterologous transgenes sequenced, allowing identification of the secreted protein. We typically also confirm the localization of candidates of interest by transient expression as GFP fusion proteins in onion epidermal cells, coupled with staining using the FM-64 dye to co-stain for the plasma membrane,
| Necrosis Inducing Protein (NIP) Secretion Trap Screen |

A Nicotiana benthamiana leaf showing regions of HR indicating the presence of a secreted protein fused to NIP.
We have also developed a new technique termed the NIP secretion trap screen. Briefly, this involves ligating cDNAs in frame at the 5’ end of the DNA sequence encoding a gene termed necrosis-inducing protein (NIP) that has been truncated to remove the native secretory signal peptide. NIP was identified from the oomycete Phytophthora sojae, as a protein that induces a hypersensitive response (HR), resulting in cell death, in leaves of some plants when present in the apoplast. It does not cause HR when expressed in the plant cytosol. Constructs containing individual cDNA::Nip fusions are transformed into Agrobacterium tumefaciens, which is then infiltrated into the apoplastic compartment of Nicotiana benthamiana leaves. Any proteins encoding SPs will be secreted from infected plant cells as NIP fusion proteins, causing a readily detectable HR.
The Nip fusion assay is a convenient and rapid means to confirm secretion of proteins in planta and complements the YST screen.
| Objectives |
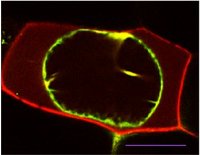
A cytoplasmic protein (green) and a secreted protein (red) in plasmolyzed onion cells. Scale bar represents 50 microns.
- Screen a range of tomato tissues for genes encoding secreted proteins, focusing particularly on fruit development and leaves at various stages of infections with Phytopthora infestans.
- Annotate the corresponding genes and confirm their localization in the cell wall by transient expression as fluorescent fusion proteins in onion epidermal cells.
- Further examine the localization and trafficking pathways of candidates that show unexpected localization in the apoplast.
To date we have identified both known cell wall localized proteins as well as a number of proteins whose presence in the apoplast is unexpected, based on functional annotation, or the absence of a predicted secretory signal peptide, suggesting alternative non-canonical secretion pathways, as have describe in yeast and mammalian cells. We have also identified several proteins that show dual localization in the wall an intracellular compartments, and we are now characterizing some of the complexity in the secretory pathway.
| Data Sets |
| Publications |

- Lee, S.J., Kim, B.-D. and Rose, J.K.C. (2006) Identification of eukaryotic secreted and cell surface proteins using the yeast secretion trap screen. Nature Protocols 1: 2439-2447.
- Lee, S.-J. Kelley, B., Damasceno, C.M.B., St. John, B., Kim, B.-S. Kim, B.-D. and Rose, J.K.C. (2006) A functional screen to characterize the secretomes of eukaryotic phytopathogens and their hosts in planta. Molecular Plant Microbe Interactions 12: 1368-1377.
- Isaacson, T., Saravanan, R.S., He, Y., Damasceno, C.M.B., Català, C., Saladié, M. and Rose, J.K.C. (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nature Protocols 1: 769-774.
- Tyler, B.M. et al. (2006) Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313: 1261-1266.
- Mueller, L.A. et al. (2009) A snapshot of the emerging tomato genome sequence. The Plant Genome 2: 78-92.
- Rose, J.K.C. and Lee, S.-J. (2010) Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiology 153: 433-436.
- McCann, M.C. and Rose, J.K.C. (2010) Blueprints for building plant cell walls. Plant Physiology 153: 365.
- Yeom, S.-I., Baek, H.-K., Oh, S.-K., Kang, W.-H., Lee, S.-J., Lee, J.M., Seo, E., Rose, J.K.C., Kim, B.-D. and Choi, D. (2011) Title: Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Molecular Plant Microbe Interactions 24: 671-684.
- Lee, S.-J. and Rose, J.K.C. (2011) Characterization of the plant cell wall proteome using high throughput screens. In The Plant Cell Wall. Methods and Protocols. Methods in Molecular Biology. 715: 255-272.
- Lee, S.J. and Rose, J.K.C. (2012) A yeast secretion trap assay for identification of secreted proteins from eukaryotic phytopathogens and their plant hosts. Methods in Molecular Biology 835:519-30.