The Tomato Fruit Glycoproteome
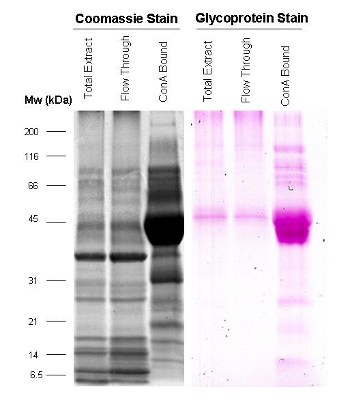
SDS PAGE gel image showing tomato fruit proteins isolated by lectin affinity, stained with Coomassie Blue (black) or glycoprotein stain (pink).
As with all eukaryotes, N-glycosylation in plant cells is a common post-translational modification for proteins traveling through the secretory pathway, many of which are destined for the cell wall. The large-scale isolation and analysis of glycoproteins by lectin affinity chromatography, coupled with mass spectrometry (MS), has become a powerful tool to evaluate the glycoproteome of mammalian cells; however, is has rarely been used with plants glycoproteins.
We are using affinity selection of glycoproteins using the lectin Concanavalin A, coupled with two-dimensional liquid chromatography (2-D LC) and matrix-assisted laser desorption/ionization mass spectrometry (2DLC-MALDI MS/MS) and electrospray ionization MS (ESI-MS) analysis, to identify extracellular and secretory pathway proteins, focusing on those expressed in ripe tomato fruit.
Tomato has long served as model system for fleshy fruit development and ripening is an excellent system to study cell wall proteins as it is associated with dramatic changes in wall biology, including dramatic enzyme-mediated cell wall polysaccharide degradation, apoplastic sugar metabolism and extracellular defenses against microbial pathogens. In spite of this, there are few published proteomic analyses of ripening tomato fruit and most of those are based on extraction of total proteins followed by 2-DE separation, since the wall proteome was not the major target. Consequently, the protein extraction step was not optimized for secreted proteins.
| Objectives |
- Develop lectin affinity chromatography protocols to optimize extraction of plant glycoproteins.
- Evaluate various MS strategies for effective identification of glycoproteins, including MALDI and ESI associated strategies.
- Generate a catalog of annotated tomato glycoproteins, focusing on fruit development, including identification of glycosylation sites.
- Confirm localization using transient expression as fluorescent fusion proteins in onion epidermal cells.
Computational prediction of the subcellular location of the N-glycoproteins isolated from green mature tomato fruit with Concanavalin A.
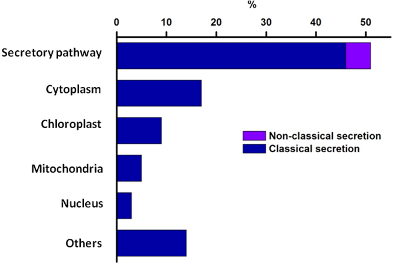
This enrichment strategy has been highly effective, with a high proportion the identified proteins showing predicted localization either in the secretory pathway. We are currently investigating the targeting of proteins from the secretory pathway to other intracellular organelles.
| Data Sets |
- Tomato (cv. Ailsa Craig) ripe fruit: annotated glycoproteins, MALDI
- Tomato (cv. Ailsa Craig) ripe fruit: glycoproteins MALDI, peptides
- Tomato (cv. Ailsa Craig) mature green fruit: glycoproteins, ESI
- Glycoprotein fractions from mature green tomato fruit identified using Concanavalin A
- Glycoprotein fractions from mature green tomato fruit identified using Snowdrop lectin
- Glycoprotein fractions from mature green tomato fruit identified using Lentil lectin
- More files in FTP
| Publications |

- Catala, C., Howe, K.J., Hucko, S., Rose, J.K.C. and Thannhauser, T.W. (2011) Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics 11: 1530-1544.
- Ruiz-May, E., Kim, S.J., Brandizzi, F. and Rose, J.K.C. (2012) The secreted plant N-glycoproteome and associated secretory pathways. Frontiers in Plant Physiology 3: 117.
- Zhang, S., Sherwood, R.W., Yang, Y., Tara Fish, T., Chen, W., McCardle, J. A., Jones, R.M., Yusibov, V., Ruiz-May, E., Rose, J.K.C. and Thannhauser, T.W. (2012) Comparative characterization of the glycosylation profiles of an influenza hemagglutinin produced in plant and insect hosts. Proteomics 12: 1269-88.
- Ruiz-May, E., Thannhauser, T.W., Zhang, S. and Rose, J.K.C. (2012) Analytical technologies for identification and characterization of the plant N-glycoproteome. In press, Frontiers in Plant Physiology.