Mass Spectrometry-based Proteomics Workshop
Mass Spectrometry-based Proteomics Workshop
April 3rd & 4th, 2012
Supported by NSF Plant Genome Program
Instruction: Drs. Jocelyn Rose and Ted Thannhauser and Sheng Zhang
Location: Cornell Proteomics & Mass Spectrometry Core Facility
We had another extremely successful Proteomics Workshop this year that was highly oversubscribed. The workshop comprised both lectures and practical lab-based training in proteomic analysis, using advanced mass spectrometry techniques and bioinformatics and was targeted towards undergraduate and graduate students. Based on the positive and enthusiastic feedback, a similar course will be offered next year.
Handouts from the workshop [pdf]
Photos from this year's workshop

April 3rd:
8:00am - 8:30am: Registration (badge, program handouts, breakfast) in room 130
8:30am - 8:35am in room 130:
- Program introduction by Professor Joss Rose
8:35am - 10:00am in room 141:
- Experimental 1: Protein denaturing, reduction and alkylation of crude cell extract samples for shotgun proteomics analysis. (Attendees divided into 4 groups)
- Instructors: Huiming Yan (bench 1), Yong Yang (bench 2), Bob Sherwood (bench 3) and Celeste Ptak (bench 4).
10:20am - 11:20am in room 130
- Lecture 1 Presentation: Sample preparation and separation prior to MS analysis by Dr. Ted Thannhauser
11:30am - 12:00pm in room 141:
- Experimental 2: in solution trypsin digestion.
12:00pm - 1:00pm in room 130: Lunch and Break
1:00pm - 2:15pm in room 130:
- Lecture 2 Presentation: MS instrument fundamentals and Applications: (proteomics case studies) by Dr. Sheng Zhang
2:30pm - 3:30pm in room 143:
- Experimental 3: Lab tour and demo, split into 3 groups (20 min/rotation)
- nanoLC-MS/MS for iTRAQ samples in 4000 Q Trap: by Dr. Celeste Ptak
- MALDI-TOF/TOF analysis for 2D gel-based protein IDs: by Bob Sherwood
- NanoLC-MS/MS analysis for 1D gel bands: by Kevin Howe
4:00pm - 5:00pm:
- Experimental 4: set up nanoLC-MS/MS overnight running for in-solution digested samples with 60-min gradient for each run.
- 10 samples on 4000 Q Trap by Dr. Celeste Ptak
- 10 samples on Synapt HDMS by Kevin Howe and Yong Yang
April 4th:
8:00am - 8:30am: breakfast and laptop computer setup in room 130
8:30am - 9:00am:
- Experimental 5: Database searching and file conversion for overnight LC-MS/MS files
- Mascot Maemon search and wiff file conversion to TMP file by Dr. Celeste Ptak in room 130
- PLGS 2.4-Mascot database search and generating PKL file by Yong Yang and Kevin Howe in room 143
9:30am - 10:30am:
- Lecture 3 Presentation: Introduction to Mascot-based MS database search and data interpretation by Professor Joss Rose
10:30am - 10:45am: transfer PKL files and TMP files into individual laptop computers.
10:45am - 11:15am: in room 130 and room 143
- Experimental 6: Practice for generating PKL files and TMP files. Instructors: Celeste Ptak for TMP files, Huiming Yan and Kevin Howe for PKL files.
12:00pm - 1:00pm in room 130: Lunch and Break
1:00pm - 3:00pm in room 130
- Experimental 7: database search practice against either Mascot public search engine or in-house licensed search engine. Assistants: Celeste Ptak, Huiming Yan, Kevin Howe and Yong Yang.
3:20pm - 4:00pm: in room 130
- Summary of the group results for shotgun analysis in 4000 Q Trap and Synapt HDMS. Summary of the iTRAQ demo results for quantitative proteomics by Sheng Zhang.
4:00pm - 5:00pm: in room 130
- Wrap-up comments, questions and suggestions

Participants in the 2012 workshop
Here are some pictures from previous workshops. Thank you all for a great time!!!

The Class of 2009... our brave band
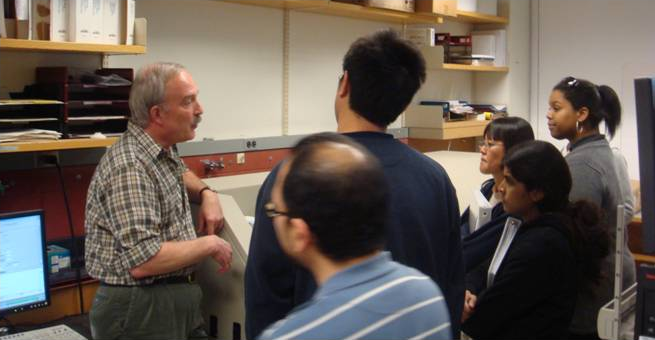
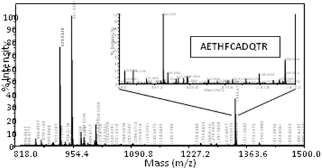
Bob explaining the mysteries of the MALDI TOF-TOF

Celeste and the art of protein fractionation

Kevin revealing some of the principles and wizardry of mass spectrometry

Rapt attention as Bob demonstrates 2D gel analysis software